之前在總機轉圖中,有給大家看過這個藥物fosfomycin,
現在找一些更詳細的機轉給大家參考:
|
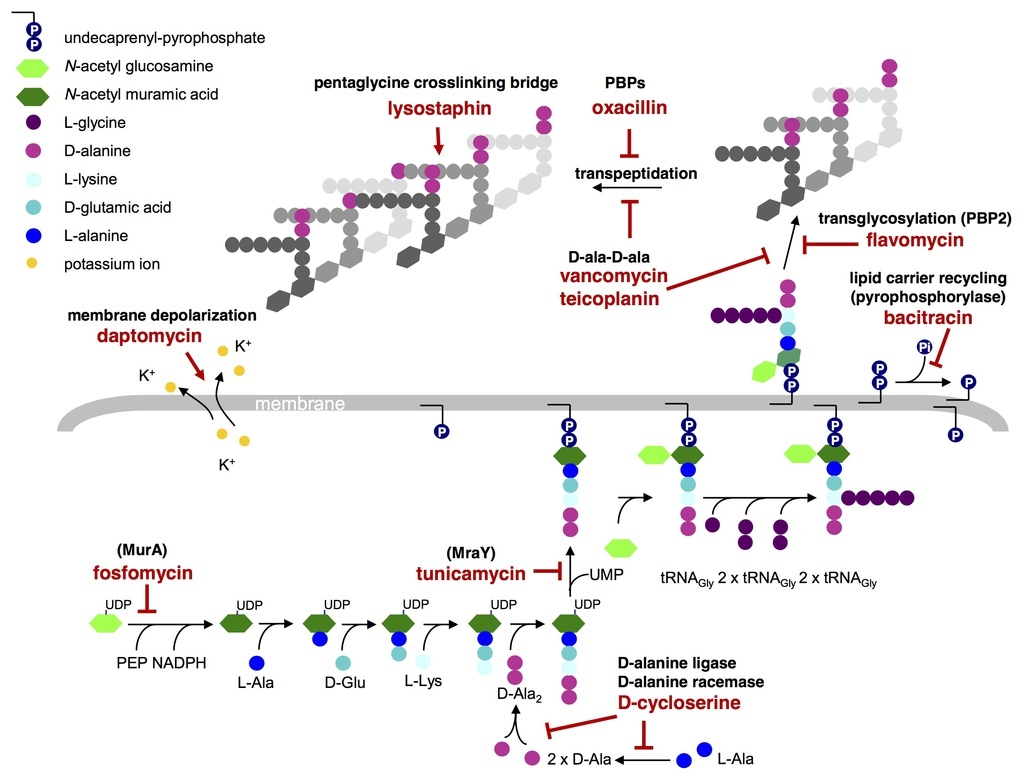
Schematic representation of the enzymatic steps involved in S. aureus cell wall synthesis and the targets of cell wall active antibiotics. Fosfomycin inhibits the enzyme MurA (UDP-N-acetylglucosamine-3-enolpyruvyl transferase) that catalyses the addition of phosphoenolpyruvate (PEP) to UDP-N-acetyl-glucosamine (GlcNAc) to form UDP-N-acetyl-muramic acid (UDP-MurNAc) . D-cycloserine prevents the addition of D-alanine to the peptidoglycan precursor by inhibiting D-alanine:D-alanine ligase A and alanine racemase . Tunicamycin is a glycoprotein antibiotic that inhibits the transfer of peptidoglycan precursor (phospho-MurNAc-pentapeptide) to the lipid carrier undecaprenyl pyrophosphate (or C55-isoprenyl pyrophosphate), catalysed by the translocase MraY Sub-lethal concentrations of tunicamycin also inhibit TarO, the first enzyme in the wall teichoic acid pathway . Bacitracin forms a metal-dependent complex with the lipid carrier undecaprenyl pyrophosphate, thereby preventing dephosphorylation and the r.ecycling of the lipid carrier required for cell wall synthesis . Flavomycin (a moenomycin complex) is a phosphoglycolipid antibiotic that inhibits transglycosylation through binding of the transglycosylase domain of penicillin-binding protein 2 (PBP2) . Glycopeptide antibiotics, such as vancomycin and teicoplanin, inhibit cell wall synthesis by binding the D-ala-D-ala of the lipid II and sterically hindering transglycosylation and transpeptidation. Teicoplanin activity is enhanced through its interaction with the cytoplasmic membrane. ß-lactam antibiotics, such as oxacillin, bind the transpeptidase active domain of penicillin-binding proteins (PBPs) by mimicking the D-ala-D-ala end of the pentapeptide . The mode of action of daptomycin is not fully known, it causes calcium-dependent disruption of membrane function and potassium efflux , but was also predicted to directly or indirectly inhibit peptidoglycan systhesis [9]. Lysostaphin is a zinc metalloenzyme that cleaves the pentaglycine crosslinking bridge specific for the cell wall of S. aureus . (Adapted from McCallum N, Berger-Bachi B, Senn MM: Regulation of antibiotic resistance in Staphylococcus aureus.Int J Med Microbiol 2010, 300(2-3):118-129.).
Dengler et al. BMC Microbiology 2011 11:16 doi:10.1186/1471-2180-11-16 |
雖然上面那張已經很清楚了,不過我還是多找一些給各位:
這是Nature chemical biology 的
August 2013, Volume 9 No 8 pp468-526
這是
Antibiotics 2013, 2, 217-236; doi:10.3390/antibiotics2020217
[Review]Molecular Mechanisms and Clinical Impact of Acquired and Intrinsic Fosfomycin Resistance
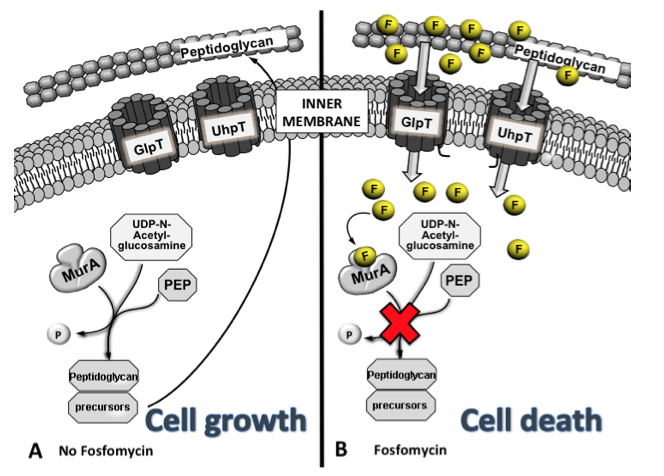
Figure 2. Although transporters are usually very selective, the chemical structure of fosfomycin mimics both glycerol-3-P (G3P) and glucose-6-P (G6P), which are transported under normal conditions. MurA catalyses the formation of UDP-GlcNac-3-O-enolpyruvate, a peptidoglycan precursor, from UDP-GlcNAc and PEP during the first step of peptidoglycan biosynthesis, allowing cell growth (A). In contrast, when fosfomycin (F) is present, it is transported inside the cell by GlpT and UhpT, blocking the UDP-GlcNac-3-O-enolpyruvate synthesis by mimicking the original substrate of MurA, PEP, avoiding cell wall synthesis and leading to cell death (B). For simplicity, only peptidoglycan and the inner membrane are shown.
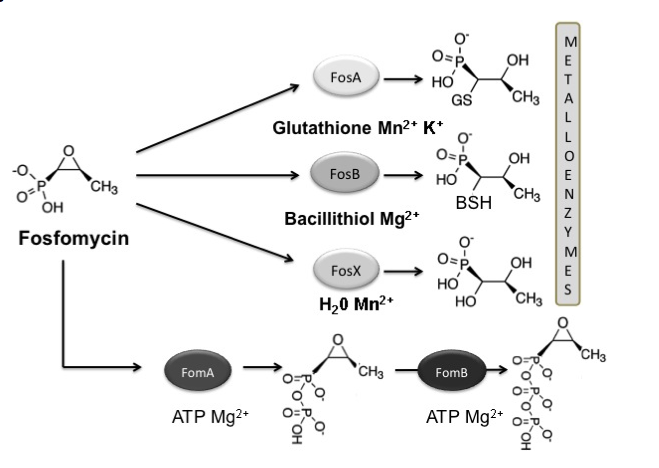
Figure 5. Reactions catalysed by Fos metalloenzymes (FosA, FosB and FosX) and fosfomycin kinases (FosC, FomA and FomB). Fosfomycin-inactivating enzymes modify the antibiotic, rendering it inactive by opening the oxirane ring (metalloenzymes) or by phosphorylation (fosfomycin kinases). Substrates and the metal requirement for each enzyme are also shown.





 留言列表
留言列表
 線上藥物查詢
線上藥物查詢 